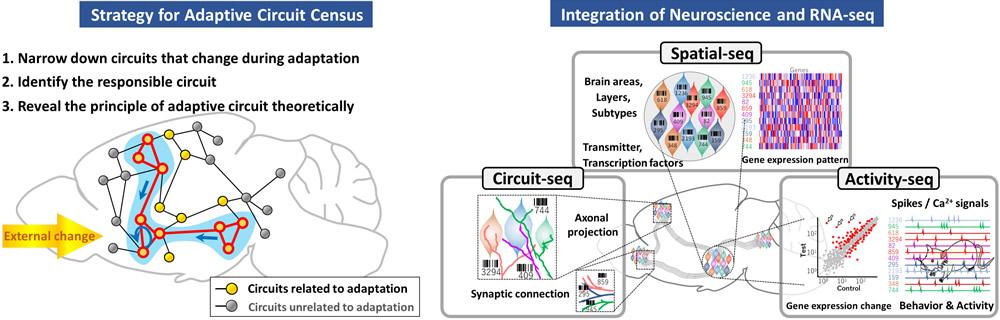
In this project area, we will focus on the circuit construction and transitions responsible for adaptive brain functions. Combining advanced neuroscience techniques for measuring and manipulating neural circuit activity with cutting-edge techniques for single-cell gene expression analysis will provide detailed information about cell type–specific adaptive circuits (Adaptive Circuit Census: ACC). Based on the adaptive circuit census, we will experimentally validate the responsible circuits and theoretically establish their operating principles.
To further promote the ACC project area, we have established a seamless, interdisciplinary cooperative framework to exchange creative and innovative ideas as well as cutting-edge experimental and analytical techniques. The project subareas are divided into A01, "Census of adaptive circuit construction," and B01, "Census of adaptive circuit transition," based on the differences of the timescale. In addition, the project subarea C01, "Technology and theory for adaptive circuit census," interacts with A01 and B01 to facilitate targeting of adaptive circuits (Research organization chart).
The A01 project subarea is organized by the Horie, Shimogori, and Fujiyama teams; the B01 project subarea is organized by the Isomura, Sasaki, and Kobayashi teams; and the C01 project subarea is organized by the Go and Shimazaki teams. In addition, publicly invited research teams will participate in each project subarea in the first and second terms of the project area. Together with the A01, B01, and C01 teams, publicly invited research teams will provide interdisciplinary approaches to study various adaptive brain functions in different species and stimulate this novel research area.
ACC (project area)
Project Leader: Yoshikazu Isomura
A01
"Cell type census for adaptive circuit construction"
Cell type census for circuit development

Team leader
Tomomi Shimogori
Team Leader
RIKEN Center for Brain Science

Co-investigator
Ai Nakashima
Associate Professor
Graduate School of Pharmaceutical Sciences, University of Tokyo
Mechanism for building diversity of neural projections responsible for behavioral selection

Team leader
Fumino Fujiyama
Professor
Faculty of Medicine and Graduate School of Medicine, Hokkaido University

Co-investigator
Nobuhiko Ohno
Professor
Department of Anatomy, Division of Histology and Cell Biology, Jichi Medical University
B01
" Cell type census for adaptive circuit transition"
Adaptive circuit dynamics for operant learning of optimal behavior

Team leader
Yoshikazu Isomura
Professor
Graduate School of Medical and Dental Sciences, Institute of Science Tokyo

Co-investigator
Nobuya Sato
Professor
School of Humanities, Kwansei Gakuin University

Co-investigator
Fuyuki Karube
Assistant Professor
Graduate School of Medicine, Hokkaido University

Co-investigator
Yutaka Satou
Associate Professor
Department of Zoology, Graduate School of Science, Kyoto University

Co-investigator
Haruka Ozaki
Associate Professor
Faculty of Medicine, University of Tsukuba
Neuronal circuit dynamics and gene expression profiles related to memory states

Team leader
Takuya Sasaki
Professor
Graduate School of Pharmaceutical Sciences, Tohoku University

Co-investigator
Akihiro Funamizu
Lecturer
Institute for Quantitative Biosciences, The University of Tokyo
Gene and cell function dynamics underlying behavioral adaptation in response to environments

Team leader
Kazuto Kobayashi
Professor
Institutes of Biomedical Sciences
Fukushima Medical University School of Medicine

Co-investigator
Natsuki Matsushita
Associate Professor
Division of Laboratory Animal Research, Aichi Medical University School of Medicine

Co-investigator
Susumu Setogawa
Lecturer
Department of Physiology, Osaka Metropolitan University Graduate School of Medicine
C01/02
"Technology and theory for adaptive circuit census"
C01 Development of comprehensive cell type census technology for adaptive neuronal circuits

Team leader
Yasuhiro Go
Professor
The Graduate School of Information Science, University of Hyogo

Co-investigator
Itoshi Nikaido
Team Leader
Laboratory for Bioinformatics Research
RIKEN Center for Biosystems Dynamics Research
C02 Theoretical approaches to adaptive neuronal circuits identified by the cell type census

Team leader
Hideaki Shimazaki
Associate Professor
Graduate School of Informatics, Kyoto University

Co-investigator
Takuma Tanaka
Professor
Graduate School of Data Science, Shiga University
Publicly invited research teams (FY2024-2025)
A01
Identification of hub networks for fear memory by combining optical and machine-learning-based approaches with comprehensive molecular profiling methods.

Identification of hub networks for fear memory by combining optical and machine-learning-based approaches with comprehensive molecular profiling methods.
Masakazu Agetsuma
Senior Researcher
Institute for Quantum Life Science, National Institutes for Quantum Science and Technology (QST)
Adaptive circuit diversity causing natural variation in temperature acclimation

Adaptive circuit diversity causing natural variation in temperature acclimation
Atsushi Kuhara
Professor
Konan University, Faculty of Science and Engineering / Institute for Integrative Neurobiology
Exploring the molecular basis of sexual dimorphism and evolutionary processes in the brain during visual partner selection

Exploring the molecular basis of sexual dimorphism and evolutionary processes in the brain during visual partner selection
Hideaki Takeuchi
Professor
Graduate School of Life Sciences, Tohoku University
Dissecting the adaptive circuit construction mechanisms by co-profiling of neuronal differentiation state and transcriptome

Dissecting the adaptive circuit construction mechanisms by co-profiling of neuronal differentiation state and transcriptome
Naoki Nakagawa
Assistant professor
National Institute of Genetics, Laboratory of Mammalian Neural Circuits
Investigating the neural circuits involved in environmental adaptation in sessile cnidarian coral using a combination of in vivo and in vitro approaches

Investigating the neural circuits involved in environmental adaptation in sessile cnidarian coral using a combination of in vivo and in vitro approaches
Koki Nishitsuji
Associate Professor
Department of Marine Science and Technology, Fukui Prefectural University
Neural mechanism underlying sex differences in sleep

Neural mechanism underlying sex differences in sleep
Hiromasa Funato
Visiting Professor
International Institute for Integrative Sleep Medicine, University of Tsukuba
Construction and Transition of Cell Assembly by Neuronal Individuality Census

Construction and Transition of Cell Assembly by Neuronal Individuality Census
Takeshi Yagi
Professor
Graduate School of Frontier Biosciences
The University of Osaka
B01
Understanding Roles of Molecular Signatures in Transition of Fear Memory Circuits in the Hippocampus by Fear Memory Engram Census

Understanding Roles of Molecular Signatures in Transition of Fear Memory Circuits in the Hippocampus by Fear Memory Engram Census
Satoshi Kida
Professor
Graduate School of Agriculture and Life Sciences, The University of Tokyo
Social circuit transition mechanisms in autism

Social circuit transition mechanisms in autism
Toru Takumi
Professor
Kobe University School of Medicine
Novel Adaptive Circuit Derived from Neural Reorganization and Impairment Recovery Models via Synthetic Synapse Connectors

Novel Adaptive Circuit Derived from Neural Reorganization and Impairment Recovery Models via Synthetic Synapse Connectors
Kosei Takeuchi
Professor
Aichi Medical University, School of Medicine
Exploring the diversity of activity-dependent translatome dynamics

Exploring the diversity of activity-dependent translatome dynamics
Hiromu Tanimoto
Professor
Graduate School of Life Sciences, Tohoku University
Census analyses of neural circuits that adaptively generate distinct behaviors

Census analyses of neural circuits that adaptively generate distinct behaviors
Akinao Nose
Professor
Graduate School of Frontier Sciences, The University of Tokyo
Local circuit census by retro-barcode

Local circuit census by retro-barcode
Kosuke Hamaguchi
Professor
Graduate School of Medical and Dental Sciences at the Kagoshima University
Transcriptome/synaptome analysis of single-cell adaptive circuit remodeling in the brain in vivo

Transcriptome/synaptome analysis of single-cell adaptive circuit remodeling in the brain in vivo
Takayasu Mikuni
Professor
Brain Research Institute, Niigata University
C01
Elucidation of brain transformation dynamics via transcription factor activity census

Elucidation of brain transformation dynamics via transcription factor activity census
Kentaro Abe
Professor
Graduate School of Life Sciences, Tohoku University
Comprehensive epigenomic analysis by a novel method for isolating specific types of neurons using FISH

Comprehensive epigenomic analysis by a novel method for isolating specific types of neurons using FISH
Yusuke Kishi
Associate Professor
The University of Tokyo, Institute for Quantitative Biosciences
Development of single-cell spatial epigenomics technology for adaptive circuit census

Development of single-cell spatial epigenomics technology for adaptive circuit census
Tetsuro Komatsu
Lecturer
Gunma University, Institute for Molecular and Cellular Regulation
Development of highly efficient trans-synaptic AAV capsid mutants

Development of highly efficient trans-synaptic AAV capsid mutants
Ayumu Konno
Lecturer
Gunma University, Gunma University Graduate School of Medicine
Spatio-Temporal Proteomics Decipher Molecular Mechanisms of Adaptive Neural Circuits

Spatio-Temporal Proteomics Decipher Molecular Mechanisms of Adaptive Neural Circuits
Tetsuya Takano
JST・PRESTO
Associate Professor, Division of Molecular Systems for Brain Function, Kyushu University Institute for Advanced Study, Medical Institute of Bioregulation
Chromatin Census Based on Neural Circuit Imaging

Chromatin Census Based on Neural Circuit Imaging
Mizuki Honda
Assistant Professor
Hiroshima University, Graduate School of Integrated Sciences for Life
C02
Distilling Adaptive Dynamics from Neuronal Populations Composed of Diverse Dynamical Information Processing Units

Distilling Adaptive Dynamics from Neuronal Populations Composed of Diverse Dynamical Information Processing Units
Yasuhiro Tsubo
Professor
College of Information Science and Engineering Ritsumeikan University
Publicly invited research teams (FY2022-2023)
A01
Identification of hub networks for fear memory by combining optical and machine-learning-based approaches with comprehensive molecular profiling methods.

Identification of hub networks for fear memory by combining optical and machine-learning-based approaches with comprehensive molecular profiling methods.
Masakazu Agetsuma
Senior Researcher
Institute for Quantum Life Science
Mechanisms of neural circuit formation in a mouse model of autism

Mechanisms of neural circuit formation in a mouse model of autism
Atsuki Kawamura
Assistant professor
Graduate School of Medical Sciences, Kanazawa University
Circuit selection and function building census in animal temperature acclimatization

Circuit selection and function building census in animal temperature acclimatization
Atsushi Kuhara
Professor
Graduate school of Natural Science & Institute for Integrative Neurobiology, Konan University
Unraveling the Molecular/Neural Underpinnings of Sexual Dimorphism in Mating Strategies Using 3D Modality Analysis Techniques

Unraveling the Molecular/Neural Underpinnings of Sexual Dimorphism in Mating Strategies Using 3D Modality Analysis Techniques
Hideaki Takeuchi
Professor
Graduate School of Life Sciences, Tohoku University
Dissecting the mechanisms underlying adaptive circuit construction by profiling the inter-neuronal heterogeneity of differentiation

Dissecting the mechanisms underlying adaptive circuit construction by profiling the inter-neuronal heterogeneity of differentiation
Naoki Nakagawa
Assistant professor
National Institute of Genetics
Understanding of the adaptive neural circuit of sedentary cnidarian coral living in various environment

Understanding of the adaptive neural circuit of sedentary cnidarian coral living in various environment
Koki Nishitsuji
Associate Professor
Department of Marine Science and Technology, Fukui Prefectural University
Complex circuit formation and cell assembly formation and transition by neuronal individuality census

Complex circuit formation and cell assembly formation and transition by neuronal individuality census
Takeshi Yagi
Professor
Graduate School of Frontier Biosciences
Osaka University
B01
Neural Circuit Mechanisms of Decision Making Based on Active Inference

Neural Circuit Mechanisms of Decision Making Based on Active Inference
Hitoshi Okamoto
Team Leader
RIKEN Center for Brain Science
Neural census of adaptive circuit shift for proactive and trial-and-error strategy

Neural census of adaptive circuit shift for proactive and trial-and-error strategy
Kei Oyama
Senior Researcher
Department of Functional Brain Imaging, National Institutes for Quantum Science and Technology
Understanding Molecular Basis of Circuit Transition from Reconsolidation to Extinction by Fear Memory Engram Census

Understanding Molecular Basis of Circuit Transition from Reconsolidation to Extinction by Fear Memory Engram Census
Satoshi Kida
Professor
Graduate School of Agriculture and Life Sciences, The University of Tokyo
Dynamics of synaptic memory circuit

Dynamics of synaptic memory circuit
Akihiro Goto
Program-Specific Associate professor
Kyoto University Graduate School of Medicine, Department of Pharmacology
Mapping of adaptive neuronal circuits for developmental song learning in zebra finches

Mapping of adaptive neuronal circuits for developmental song learning in zebra finches
Yoko Yazaki-Sugiyama
Associate Professor
Neuronal Mechanism for Critical Period Unit, OIST Graduate University
Mechanism of circuit adaptation through translatome dynamics

Mechanism of circuit adaptation through translatome dynamics
Hiromu Tanimoto
Professor
Graduate School of Life Sciences, Tohoku University
Modification mechanism of neural circuit dynamics responsible for memory transition by neuropeptides

Modification mechanism of neural circuit dynamics responsible for memory transition by neuropeptides
Ayako Tonoki
Associate Professor
Graduate School of Pharmaceutical Sciences, Chiba University
Census analyses of neural circuits that adaptively generate distinct behaviors

Census analyses of neural circuits that adaptively generate distinct behaviors
Akinao Nose
Professor
Graduate School of Frontier Sciences, The University of Tokyo
Census of the functional neuron activity in the fronto-striatal circuit

Census of the functional neuron activity in the fronto-striatal circuit
Satoshi Nonomura
Lecturer
Shiga University of Medical Science
Local Circuit Analysis with Retrograde Barcodes

Local Circuit Analysis with Retrograde Barcodes
Kosuke Hamaguchi
Associate Professor
Graduate School of Medicine, Kyoto University
Spatiotemporal mechanisms for the adaptive circuit rearrangement in single neurons in vivo

Spatiotemporal mechanisms for the adaptive circuit rearrangement in single neurons in vivo
Takayasu Mikuni
Professor
Brain Research Institute, Niigata University
C01
Transcription factor activity census for elucidating mechanism behind brain plasticity

Transcription factor activity census for elucidating mechanism behind brain plasticity
Kentaro Abe
Professor
Graduate School of Life Sciences, Tohoku University
Simultaneous voltage and calcium imaging and mRNA extraction from single neurons in-vivo

Simultaneous voltage and calcium imaging and mRNA extraction from single neurons in-vivo
Christopher Roome
Staff Scientist
Okinawa Institute of Science and Technology Graduate University (OIST)
C02
Extracting Adaptive Neural Networks through Spike Statistical Techniques for Heterogeneous Neural Populations

Extracting Adaptive Neural Networks through Spike Statistical Techniques for Heterogeneous Neural Populations
Yasuhiro Tsubo
Professor
College of Information Science and Engineering Ritsumeikan University
A seamless neuroscience research group, such as one that combines single-cell gene analysis techniques and circuit manipulation, has not been effectively created in Japan. Therefore, we established the research support committee (histological analysis, physiological analysis, behavioral analysis, gene manipulation, and theoretical/statistical analysis support teams) and transcriptomic analysis support committee (collaboration coordination and RNA-seq analysis unit) within the executive committee of the Administrative organization (Admin organization chart).
In addition to systematic technological support, we provide a variety of support to stimulate scientific activities for ACC researchers, including academic exchanges, international moves, fostering of young researchers, and contributions to society. Overall, we aim to understand the true "Adaptive Circuit Census" by obtaining valuable research findings using innovative technologies, thus providing a seamless collaboration platform for the next generation of young researchers.
| Executive committee | Yoshikazu Isomura, Takeo Horie, Tomomi Shimogori, Fumino Fujiyama, Takuya Sasaki, Kazuto Kobayashi, Yasuhiro Go, Hideaki Shimazaki |
| Research support committee | Kobayashi (chair)Histological analysis support unitFujiyama Physiological analysis support unitSasaki, Funamizu, Isomura Behavioral analysis support unitSato (n) Gene manipulation support unitKobayashi, Matsushita Theoretical/statistical analysis support unitShimazaki, Ozaki, Tanaka |
| Transcriptomic analysis support committee | Go (chair)Collaboration coordination unitGo, Horie RNA-seq analysis unitGo, Horie, Ozaki, Nikaido |
| Young researcher support committee Global activity stimulation committee Conference planning committee Outreach committee External evaluation committee ACC office | Sasaki Shimogori Horie Fujiyama Masahiko Watanabe (Hokkaido Univ.), Masanobu Kano (Tokyo Univ.), Noriyuki Sato (OIST), Nenad Sestan (Yale Univ.) Isomura |
* Horie had participated until August 2024.